How To Get the Best Out of ‘R’ Programming

R is one of the most powerful and widely used programming languages for statistical computing. In recent years, ‘R’ has made substantial progress with Big Data and Machine learning.
The evolving and vibrant ‘R’ community has always paced itself by providing unique packages in Big Data services that can be leveraged for storage, processing, visualization, etc. At the same time, it has tons of libraries for Machine Learning applications.
With the advent of Big Data, the complexity of processing huge amounts of data has greatly reduced. Machine Learning on this humongous data is the next logical step and inevitable. Given all the challenges related to deriving actionable insights from data and the role of Machine Learning, ‘R’ could be a great tool to achieve progress on that front.
In this article, I will detail ‘Parallel Processing’ in R and how to get the most out of ‘R’ using the same. Parallel processing in ‘R’ is an excellent aid for the holistic implementation of IOT, Big Data, and Machine Learning applications.
Parallelization in R
R, by default, works on the single core available on the machine. To execute the ‘R’ code in parallel, first, you must make a cluster with the required number of seats available to R. You can do this by registering a ‘parallel backend.’ You can use the ‘parallel’ package, which creates an R instance on each core specified. The package called ‘for each’ should then define the code that needs to be executed parallelly or sequentially.
Below is a code snippet in R For Parallelization
#Libraries used for parallel processing library(doParallel) #doParallel package helps register core and parallel backend.
Library (for each) #foreach package provides a looping construct for executing R code repeatedly.
#Detect a number of cores and make a cluster with available cores on the machine. The machine we used for this snippet has 4 cores.
cl=makeCluster(detectors())
#Registering cores for parallel backend can be tested according to resource utilization.
registerDoParallel(cl, cores = detectCores()-1)
#The for each package’s main reason is that it supports parallel and sequential execution.
# ‘%dopar%’:- Executes the code in parallel on each core.
# ‘%do%’:- Executes the code sequentially on a single core.
# .packages:- Packages required for executing code within foreach(){}.
# .combine:- Operations performed with data. Ex rbind,cbind,+,*,c()
# .export:- Exporting parent environment variables.
for each(i = 1:length(datasets),.combine=c,.export=c(list of functions to be exported to loop),.packages=c(“string”,”log4r”,”parallel”,”doParallel”,….)) %dopar%
{
// applying statistical functions, algebraic expressions, mathematical computations, etc. in data science
// batch processing
// data pre-processing techniques like cleaning, formatting, transformations, etc., before passing to ML algorithms and predictions.
//real-time and time series data processing
}
# Stop the parallel backend registered
stop cluster(cl)
I have used ‘R’ parallelization for optimizing execution time in Machine Learning, batch processing, real-time data analysis, etc., on top of Big Data. I will share my insights in detail in my next article.
Monitoring Parallel Processes In R
Parallelization in ‘R’ allows continuous monitoring of threads and instances in concurrent execution. The processes of R instances on a number of cores, RAM, and CPU utilization can be tracked by ‘System Monitor API’ or top command in Ubuntu, ‘Activity monitor API’ in Mac, and ‘Windows task manager’ in Windows.
Below is a result of an execution of the R script in parallel. When you make a cluster and register the cores, R instances are allocated on each core. You can observe 4 R processes in the processes tab below (Fig.1). The memory column shows the memory usage in MiB, which is close to MB.
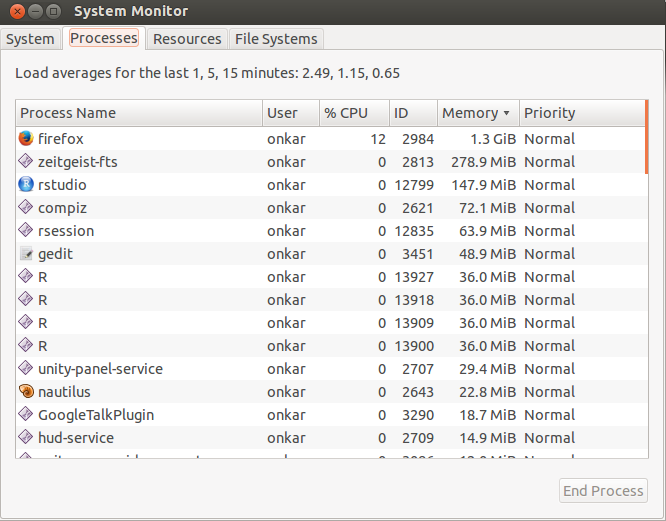
The resources tab shows the number of cores on the machine and its utilization as you can observe in the ‘CPU history’ chart, the graph spikes in 5 seconds from 20-30% to 100 %. This happens when the code for each is executed. The ‘Memory and Swap History ‘shows the total memory used and available during execution. Swap memory is used from hardisks if processes run out of RAM. If processes exceed memory and swap, the machine might freeze and require a hard boot.
The variability pattern for ‘Memory and Swap History ‘and ‘Network History’ will be observed when we schedule the R parallel process to run continuously in iterations across clusters.
In the above scenarios, you can observe the step-by-step execution across use cases. It also helps in start to end analysis of the code.
Logging
Logging for each node in each package helps in analyzing node-specific datasets. For a real-time data use case when data is received for 1000 parameters from various sources, narrowing down the reason for fatality will be required quite often and can be cumbersome. These fatalities can be device-specific, server-specific, data-specific, coding-specific etc. Processing each parameter-specific data set and logging parameter-specific details will help in faster root cause analysis.
Some of the time series data challenges, like dealing with data inconsistency, data lag, data corruption, data duplicates, data rejection/validation, and data cleaning, can be sold at a granular level. The usual method of logging in to txt files will be helpful in the initial stages when we are tweaking the application end to end(data source to the visualization), but after some time, managing these files and traversing through them becomes an activity. Bird’s eye view logging can be done in a well-designed database schema to monitor the status of real-time data and build a UI on top of it. This will give some quick data monitoring stats without manual intervention and eliminate the hassle of maintaining log files.
‘R’ Coding Tips
Here are some ‘R’ programming tips to get the best from it:
- R is memory-extensive and does a lot of in-memory processing. Hence, creating an R instance on all the cores for processing hampers resource utilization. Hence, code optimization with sequential and parallel approaches can reduce unnecessary CPU consumption. For memory-intensive processes, it is better to parallelize at a small scale in subroutines.
- Remove objects with large data sets from the workspace using rm(). If running various iterations on every core of the R instance, use rm(list=ls()) to complimentary memory from ram.
- Call garbage collector gc() wherever necessary. R is expected to do it implicitly for releasing memory, but it will not return it to the operating system. In parallel processing, each process must return memory after completion.
- Database connections need to be tracked! Hence, timely termination helps with connection pooling.
- For optimizing and avoiding loops in R, you will find many research papers to build the best out of R, and you should follow the recommendations given as a coding convention in development.
I hope you find this article helpful and get the best out of R!